| Accession | Acc. Gene-, Name | Start | End | Subsequence | Logic | PDB | Organism | Length |
|---|---|---|---|---|---|---|---|---|
| ELMI002523 | Q38825 IAA7 IAA7_ARATH |
82 | 94 | AKAQVVGWPPVRNYRKNMMT | TP |
2P1Q 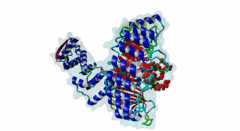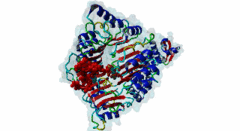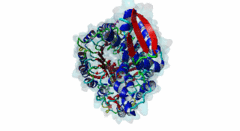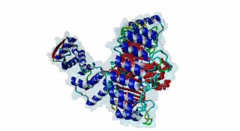
|
Arabidopsis thaliana (Thale cress) | 243 |
![]() Instance evidence
Instance evidence
| Evidence class | PSI-MI | Method | BioSource | PubMed | Logic | Reliability | Notes |
|---|---|---|---|---|---|---|---|
| experimental | MI:0096 | pull down | in vivo/in vitro | support | certain | InteractionDetection | |
| experimental | MI:0519 | glutathione s tranferase tag | in vitro | Gray,2001 | support | certain | |
| experimental | MI:0019 | coimmunoprecipitation | in vivo/in vitro | Gray,2001 | support | certain | InteractionDetection |
| experimental | MI:0074 | mutation analysis | in vivo/in vitro | Gray,2001 | support | certain | FeatureDetection |
| experimental | MI:0114 | x-ray crystallography | in vitro | Tan,2007 | support | certain | InteractionDetection FeatureDetection |
![]() Switches
Switches
This ELM instance is part of the following 1 switching mechanism annotated at the switches.ELM resource:
-
SWTI000712:
Binding of Auxin-responsive protein IAA7 (IAA7) to Protein TRANSPORT INHIBITOR RESPONSE 1 (TIR1), an F-box substrate recognition subunit of the SCF E3 ubiquitin ligase, depends on binding of the plant hormone 22676" target="_blank">auxin to TIR1, as this pre-assembled complex provides a composite binding site for the degron motif in IAA7. Subsequent ubiquitylation of IAA7 by the SCF targets this transcriptional repressor for proteasomal degradation, a pre-requisite for the transcriptional response to auxin.
![]() Pathways
Pathways
Please cite:
ELM-the Eukaryotic Linear Motif resource-2024 update.
(PMID:37962385)
ELM data can be downloaded & distributed for non-commercial use according to the ELM Software License Agreement
ELM data can be downloaded & distributed for non-commercial use according to the ELM Software License Agreement

