| Accession: | |
|---|---|
| Functional site class: | PTS1 |
| Functional site description: | PTS1 is a C-terminal signal interacting with the Pex5p protein to direct proteins into the peroxisomal matrix. |
| ELMs with same tags: |
|
| ELM Description: | Generic PTS1 ELM for all eukaryotes |
| Pattern: | (.[SAPTC][KRH][LMFI]$)|([KRH][SAPTC][NTS][LMFI]$) |
| Pattern Probability: | 0.0000152 |
| Present in taxon: | Eukaryota |
| Interaction Domain: |
TPR_1 (PF00515)
Tetratricopeptide repeat
(Stochiometry: 1 : 1)
|
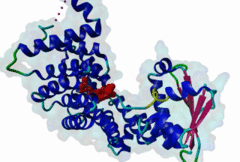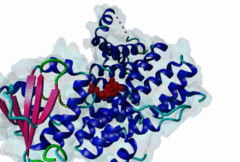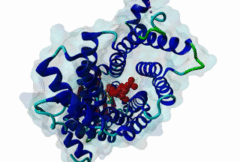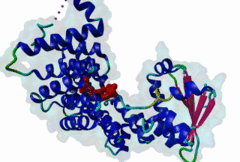
PTS1, peroxisomal targeting signal 1, is one of two peroxisomal import signals which direct proteins into the lumen (matrix) of the peroxisome, the other being PTS2. A third signal is used to target peroxisomal membrane proteins. PTS1 was first clearly defined by Gould,1989 following earlier work from the same group. Reviews of peroxisome biogenesis and import include Kunau,2001, Purdue,2001, Terlecky,2000, Holroyd,2001, Subramani,2000 and Fujiki,2000. The functions of peroxisomes vary in different cell types or organisms and some specialised peroxisomes have other names. A generic term for all these organelles is microbody. Glyoxysomes provide the glycolytic cycle in germinating plant seeds. Glycosomes perform glycolysis in trypanosomatids. Cholesterol synthesis occurs in animal peroxisomes. Two widely conserved functions are H2O2-based respiration and beta-oxidation of fatty acids. Pex5p (Peroxin 5) protein is the receptor for PTS1 (P50542). It imports folded proteins (and probably even imports pre-assembled complexes since some peroxisomal proteins lack obvious PTS signals). The import machinery is clearly complex and Pex5p interacts with several other proteins involved in peroxisome import, including Pex8p, 10p, 12p, 13p, 14p. During import, Pex5p is thought to shuttle between the cytosol and the peroxisomal matrix. Earlier names for Pex5p are Pas8 and Pxr1. A structure of the Pex5p-PTS1 complex (1FCH) has been solved (Gatto GJ,2000). Pex5p has 6 rod-forming TPR (tetratricopeptide) repeats, split 3-3, which fold at a hinge region to sandwich PTS1 in a cleft. The structure shows many details of the molecular recognition of PTS1 by Pex5p, including tight interaction with the C-terminal carboxyl group. The presence of PTS1 on a protein does not exclude alternative cellular locations, particularly in the cytoplasm or mitochondrion. For example mitochondria and peroxisomes have partially duplicated lipid biochemistry. Several proteins are targeted to both mitochondria and peroxisomes, either because of alternatively-spliced gene products or because both targeting signals are present on the same protein. Mutations in Pex5p define complementation group 2 of the peroxisome biogenesis disorders (PBDs), such as Zellweger syndrome (see OMIM:214100, OMIM:600414, OMIM:202370). These disorders are characterised by an absence of peroxisomes, multiple enzymatic deficiencies and physical malformation. The classical PTS1 motif is SKL$, with similar residues tolerated at each position. It has also been noted that PTS1s lacking the positively charged residue are then preceded by a K or R, presumably in binding affinity compensation, e.g. KANL$ is the PTS1 in human catalase. The current pattern will detect most examples in any eukaryote. There is no strong taxonomic divergence (as of 2002) although there may be scope for some future revision of the pattern. For example, Ala in the third (Leu) position has been seen in one or two yeast candidate PTS1s. See also the following PROSITE entry for PTS1: PS00342. |
(click table headers for sorting; Notes column: =Number of Switches, =Number of Interactions)
| Acc., Gene-, Name | Start | End | Subsequence | Logic | #Ev. | Organism | Notes |
|---|---|---|---|---|---|---|---|
| Q9UHK6 AMACR AMACR_HUMAN |
379 | 382 | EIYQLNSDKIIESNKVKASL | TP | 1 | Homo sapiens (Human) | |
| P22307 SCP2 NLTP_HUMAN |
544 | 547 | NMGLAMKLQNLQLQPGNAKL | TP | 1 | Homo sapiens (Human) | |
| P04040 CAT CATA_HUMAN |
524 | 527 | AIHTFVQSGSHLAAREKANL | TP | 3 | Homo sapiens (Human) | |
| P08659 Luciferin 4-m LUCI_PHOPY |
547 | 550 | DARKIREILIKAKKGGKSKL | TP | 2 | Photinus pyralis (Common eastern firefly) | |
| Q28956 17beta-estrad Q28956_PIG |
734 | 737 | RGNIMLSQKLQMILKDYAKI | TP | 2 | Sus scrofa (Pig) |
Please cite:
ELM-the Eukaryotic Linear Motif resource-2024 update.
(PMID:37962385)
ELM data can be downloaded & distributed for non-commercial use according to the ELM Software License Agreement
ELM data can be downloaded & distributed for non-commercial use according to the ELM Software License Agreement