| Accession: | |
|---|---|
| Functional site class: | NES Nuclear Export Signals |
| Functional site description: | The Nuclear Export Signal (NES) is a linear motif involved in the regulated export of macromolecules from the nucleus via the nuclear pores. Import and export of macromolecules is the main way to communicate between cytosol and nucleus. Therefore, although many nuclear proteins do not need to be re-exported, a substantial number of nuclear proteins can shuttle between the nucleus and cytoplasm. In such proteins, the accessibility of the import and export signals needs to be regulated, or they will immediately be returned to the other compartment. Thus, open and closed conformations of NES-bearing proteins are likely to be common features. The nuclear exporter protein, CRM1 mediates the nuclear export of cargo proteins by recognizing amphipathic NESes. These bind in a number of permutations including forward and reverse orientations. Mutations in the NES motif or their recognition regions can lead to several pathological conditions in the cell, including cancer. |
| ELMs with same func. site: | TRG_NES_CRM1_1 TRG_NESrev_CRM1_2 |
| ELM Description: | Accessible peptides matching the leucine-rich nuclear export signal (NES) motif bind to the CRM1 exportin protein. Since the NES has been reported as four to five conserved hydrophobic residues, it has presented a particular problem for experimental verification. Solving 3D structures of putative NES motif-bearing proteins has often revealed that the apparent export signals are buried in the hydrophobic core of globular domains where they cannot function as linear motifs (Gibson,2013). While some of the reported NES peptides are therefore unlikely to be true, many proteins do return to the cytoplasm from the nucleus by this mechanism. The NES pattern in ELM was therefore derived exclusively from reported instances that were not buried in domain cores. The conserved motif is found to be longer and less hydrophobic than originally reported, with a negatively charged residue at each end. An educational structure to view is the MapKapK2 kinase (1KWP), where the NES is conditionally packed onto the kinase domain (Meng,2002) but the negative residues are solvent exposed. There are indications that further negative charges may play a role (la Cour,2004). Solved CRM1-NES complexes (Monecke,2009; Dong,2009) show that a number of complementary positively charged residues are spaced around the NES-binding groove. The structures also show most of the NES peptide is helical when bound, indicating that proline cannot occur in some of the less conserved positions. With additional structures being solved it became clear that there are five hydrophobic pockets in CRM1. Forward orientation NESes may occupy all five pockets (P0-P4) or may only occupy four hydrophobic pockets, omitting either P0 or P4. Because of this variability, searching for NESes typically used only four hydrophobic positions such as Φ1-(x)2–3-Φ2-(x)2–3-Φ3-x-Φ4 (Lee,2019). This is the case for the current NES forward motif in ELM and therefore it may fail to discover the all-helical NES variants that would not have that spacing. |
| Pattern: | ([DEQ].{0,1}[LIM].{2,3}[LIVMF][^P]{2,3}[LMVF].[LMIV].{0,3}[DE])|([DE].{0,1}[LIM].{2,3}[LIVMF][^P]{2,3}[LMVF].[LMIV].{0,3}[DEQ]) |
| Pattern Probability: | 0.0007626 |
| Present in taxon: | Eukaryota |
| Interaction Domain: |
Xpo1 (PF08389)
Exportin 1-like protein
(Stochiometry: 1 : 1)
|
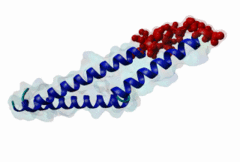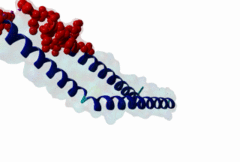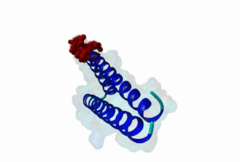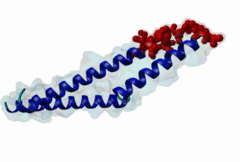
The import-export protein traffic through the nuclear envelope is mediated by soluble transport receptors (carriers) termed importins and exportins that bind to specific signals present within their substrates. The best-characterized export carrier is CRM1 (also known as exportin1/Xpo1), an evolutionarily well-conserved protein (Fornerod,1997; Wing,2022). CRM1 shuttles between the nucleus and cytoplasm binding cargo molecules in the presence of RanGTP inside the nucleus. The complexes then traverse the nuclear pores and release the cargo into the cytoplasm upon hydrolysis of the Ran-bound GTP. CRM1-mediated export is inhibited by the fungicide leptomycin B (LMB), providing a unique experimental tool for studying nuclear localization and trafficking in eukaryotic cells. LMB also aided the development of additional small molecule inhibitors of CRM1-mediated nuclear export. Although the side effects are pronounced, Selinexor was approved in 2019 in a combination drug therapy of last resort for relapsed refractory multiple myeloma (RRMM) (Richter,2020). The CRM1 mutation E571K may occur in cancer cells. This residue is present in the NES binding groove, and studies have shown that most of the NESes are not affected by this mutation, with exceptions including Mek1, eIF4E-transporter, and RPS2 that are especially highly charged (Baumhardt,2020). The NES motif was first identified in the HIV Rev protein (Fischer,1995) and in the Protein Kinase A inhibitor (PKI-alpha) (Wen,1995). Subsequently many more NESes have been identified and crystal structures have shed light on variations in how they bind to CRM1 (Wing,2022). The majority of the export substrates of CRM1 contain an amphipathic Leucine-rich Nuclear Export signal (NES) consisting of 4-5 hydrophobic residues in a region of ~10 amino acids with other residues interspersed between the key hydrophobic residues. The latter binds to a hydrophobic groove on the convex side of the CRM1 ring formed by HEAT repeats 11 and 12. The NES binding groove contains five hydrophobic pockets (P0-P4), with P0 being the shallowest. P0-P2 grooves are wider and specifically bind parts of NESes that are helical while the narrow groove beyond P3 binds either extended residues (ΦXΦ, where Φ is a hydrophobic amino acid) or a continuation of the helix. NESes adopt several variations in conformations to bind into the CRM1 groove; most bind in the classical forward direction, while certain others bind in the reverse direction (Fung,2015). Forward orientation NESes may occupy all five pockets (P0-P4) or may only occupy four hydrophobic pockets, omitting either P0 or P4. Because of this variability, searching for NESes typically used only four hydrophobic positions such as Φ1-(x)2–3-Φ2-(x)2–3-Φ3-x-Φ4 (Lee,2019). Forcing the Φ-x-Φ spacing in the pattern will probably prevent matches to the all helical NES variant. The reverse NES has a consensus pattern Φ1xΦ2xxxΦ3xxΦ4xxΦ5 where the hydrophobic residues Φ1-Φ5 bind the same pockets but in reverse order (Fung,2015). All NESes make a main chain hydrogen bond with the CRM1 Lys568 side chain that acts as a specificity filter which occludes the non-NES peptides (Fung,2017). In addition to the key hydrophobic residues, there are clear negative charge preferences in and around the NESrev core motif. NESdb is a database of experimentally reported NESes (Fung,2021). LocNES is a resource to predict NESes (Xu,2015). Genuine NES motifs appear to be always in regions of natively disordered polypeptide. Clashes with known globular domains should be assumed to indicate the non-viability of the motif candidate (Gibson,2013). However, NES motifs can be found close in sequence to known domains and may conditionally bind to them in a closed conformation. Regulated accessibility of the NES may be important for many proteins that must stay in the nucleus until signalled to exit, for instance, the MapKapK2 NES can fold back onto its kinase domain (1KWP; Meng,2002). The p53 NES is only available in the monomeric protein, being part of the tetramerization module (Foo,2007). Regulation of nuclear export may be more sophisticated than import: one indication is that CRM1 is much more flexible and dynamic (and harder to crystallize without accessory proteins such as RAN) than importins, although being composed of similar HEAT repeat elements. A standard crystallisation protocol for CRM1 in complex with NESes has been devised using the ternary complex of CRM1-Ran-RanBP1 (Fung,2022). |
(click table headers for sorting; Notes column: =Number of Switches, =Number of Interactions)
Please cite:
ELM-the Eukaryotic Linear Motif resource-2024 update.
(PMID:37962385)
ELM data can be downloaded & distributed for non-commercial use according to the ELM Software License Agreement
ELM data can be downloaded & distributed for non-commercial use according to the ELM Software License Agreement