| Accession: | |
|---|---|
| Functional site class: | TRFH domain docking motifs |
| Functional site description: | The TRF1 and TRF2 paralogues are part of a 6 protein complex termed shelterin which protects mammalian telomeres. The TRFs also recruit other proteins to the telomeres. With their TRFH domains, they not only form homodimers but also provide additional protein binding interaction surfaces. Several shelterin-associated proteins have been found to dock to a TRFH surface groove which accepts [FY]xLxP peptide motifs. |
| ELM Description: | The TRFH binding motifs are found in proteins recruited to the shelterin complex by TRF1 and TRF2. The core motif conservation is [FY]xLxP where TRF1 prefers F and TRF2 prefers Y (Chen,2008). These conserved residues make favourable contacts in a hydrophobic groove, while the peptide backbone also makes several polar contacts. In the solved TRF1-Tin2 complex, a C-terminal extension of the motif in Tin2 has positively charged residues that make favourable contacts. While it may be possible to extend the motif pattern with more data, the peptide in the TRF2-Apollo complex was shorter (and the Apollo sequence lacks equivalent charges anyway). Therefore the generic current motif excludes these interactions. Motifs are confirmed in Tin2, Apollo and PinX1. |
| Pattern: | [FY].L.P |
| Pattern Probability: | 0.0002240 |
| Present in taxon: | Eukaryota |
| Interaction Domain: |
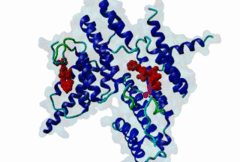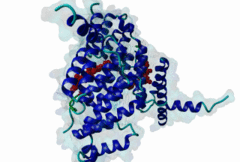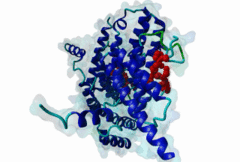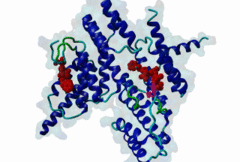
TRF (PF08558)
Telomere repeat binding factor (TRF)
(Stochiometry: 1 : 1)
|
The ends of human chromosomes are characterised by specific arrays of TTAGGG repeats at the telomeres. A complex formed by six-proteins (called Shelterin), specifically recognises these regions (Palm,2008). This complex allows cells to distinguish telomeres from sites of DNA damage. The components of shelterin were gradually identified over the past 10 years. There are essentially six core shelterin subunits: TRF1, TRF2, TIN2, RAP1, TPP1, and POT1. TRF1, TRF2 and POT1 have DNA-binding domains and directly recognize the human chromosome ends. For the shelterin-associated proteins they are recruited to telomeres through interaction with TRF1 or TRF2. TRF2 is also implicated in the regulation of DNA repair processes. TRF1 and TRF2 are paralogues with almost the same molecular architecture, yet they interact with different protein sets. They are composed of a C-terminal DNA-binding SANT domain, a dimerisation and protein-interacting TRFH domain (PF08558) and natively disordered linkers. The interaction of the shelterin protein TIN2 with TRF1 TRFH was narrowed down to an FxLxP-based motif which binds in part by anti-parallel beta augmentation (Chen,2008). A similar YxLxP motif in the shelterin accessory factor Apollo (SNM1B) binds equivalently to the TRF2 TFRH. It is thought that the motif binding pockets dictate the specificity for the F and Y residues. TRF1 also interacts with an FxLxP motif in the shelterin associated factor PinX1. |
(click table headers for sorting; Notes column: =Number of Switches, =Number of Interactions)
| Acc., Gene-, Name | Start | End | Subsequence | Logic | #Ev. | Organism | Notes |
|---|---|---|---|---|---|---|---|
| Q9H816 DCLRE1B DCR1B_HUMAN |
504 | 508 | FRGLALKYLLTPVNFFQAGY | TP | 4 | Homo sapiens (Human) | |
| Q96BK5 PINX1 PINX1_HUMAN |
289 | 293 | QPPEGRDFTLKPKKRRGKKK | TP | 2 | Homo sapiens (Human) | |
| Q9BSI4 TINF2 TINF2_HUMAN |
258 | 262 | EPLAGRHFNLAPLGRRRVQS | TP | 4 | Homo sapiens (Human) |
Please cite:
ELM-the Eukaryotic Linear Motif resource-2024 update.
(PMID:37962385)
ELM data can be downloaded & distributed for non-commercial use according to the ELM Software License Agreement
ELM data can be downloaded & distributed for non-commercial use according to the ELM Software License Agreement