| Accession: | |
|---|---|
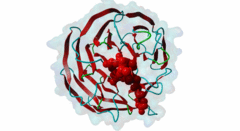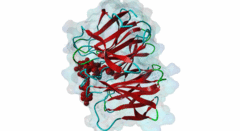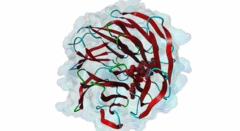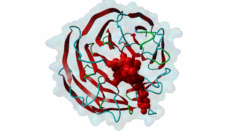
| Functional site class: | WDR5 WD40 repeat (central)-binding ligand |
| Functional site description: | In the nuclei of eukaryotic cells, DNA is complexed with histones into nucleosomes. Post-translational modification of histones regulates their interactions with DNA and other nuclear proteins, and is important for the control of cellular processes such as gene transcription, cell cycle progression and DNA repair. Important modifications include methylation of H3 histones at lysine 4 by Set1/MLL protein family members and acetylation of H4 histones at lysine 16 by MYST protein family members. Activity of these enzymes depends on their assembly in multi-protein histone modification complexes. The WD40 repeat domain protein WDR5 plays a key role in H3K4 methylation and H4K16 acetylation by acting as a scaffold protein for the assembly of the respective core histone methylation and acetylation complex, which are conserved through evolution. The recruitment of different complex subunits by WDR5 depends on distinct motifs in WDR5-binding partners, including the catalytic subunits and the accessory proteins. |
| ELMs with same tags: |
|
| ELMs with same func. site: | LIG_WD40_WDR5_WIN_1 LIG_WD40_WDR5_WIN_2 LIG_WD40_WDR5_WIN_3 |
| ELM Description: | The conserved WDR5 interaction (Win) motif was first characterized in Set1/MLL family members. Structural data show that the Win motifs in these different proteins have a similar binding mode, consistent with their sequence homology. However, notable differences could be observed, which could explain the wide range of interaction affinities displayed by the different peptides for WDR5 (Zhang,2012; Dharmarajan,2012). At its center, the Win motif contains an invariant arginine residue (position 0) that inserts into the central tunnel of the WD40 repeat domain of WDR5 (opposite to the LIG_WD40_WDR5_VDV_1 and LIG_WD40_WDR5_VDV_2 binding site). Surrounding this arginine are small residues that fit tightly at the entrance of the arginine-binding pocket. When bound, part of the peptide adopts a 3-10-helical structure that is stabilized by intra-peptide hydrogen bonds between the conserved Glu or Gln in the +2 position and the N-terminal part of the motif. One structural difference between the different Set1/MLL peptides involves residues in the +3 and +4 position. The residue in the +4 position can bind in one of two distinct pockets, called A and B, on the WDR5 surface, and the path the peptide takes appears to depend on the residue in the +3 position, with valine in the +3 position directing the residue in the +4 position to the A pocket and proline or glycine in +3 resulting in binding of the +4 residue to the B pocket. Other differences that result in the varying affinities include a water-mediated hydrogen bond in the MLL1-WDR5 interaction that is replaced by a direct hydrogen bond in the MLL4-WDR5 interaction (Zhang,2012; Dharmarajan,2012). More recently, a Win motif was found in KANSL1, a subunit of a distinct histone modification complex (Dias,2014). The main difference with the Win motif in Set1/MLL proteins is the presence of an arginine in the +2 position. |
| Pattern: | [SCA]AR[STCA][EQR][PGILVM][HYFQNKRLVI] |
| Pattern Probability: | 0.0000064 |
| Present in taxon: | Vertebrata |
| Interaction Domain: |
IPR017986 (IPR017986)
WD40-repeat-containing domain
(Stochiometry: 1 : 1)
|
Chromatin is packaged DNA in the nuclei of eukaryotic cells made up of a complex of DNA and proteins. The nucleosome units forming the higher-order chromatin structure are composed of an octamer of four highly conserved histones around which the DNA is wound. The N-terminal tails of histones undergo multiple covalent post-translational modifications in order to secure gene regulation, such as acetylation, methylation, phosphorylation, sumoylation and ubiquitylation. Incorrect histone modifications have been associated with developmental defects and different forms of cancer (Bhaumik,2007). One of the most conserved modifications is methylation of the histone H3 lysine 4 residue (H3K4), which can be mono-, di-, or trimethylated by the KMT2 family of SET domain methyltransferases. Depending on the amount of methylations, different reactions can take place. Hence, a tight regulation of the methyltransferases is essential. Six members belong to the KMT2/SET family (Set1a, Set1B and four mixed lineage leukemia (MLL) proteins) and each possesses a C-terminal conserved catalytic SET domain (PF00856). MLL1 is associated with expression of HOX genes and deregulation of the histone-modifying enzyme MLL1 has been linked to acute myeloid and lymphoblastic leukemia. In mice, a rearrangement of the MLL1 gene leads to defects in hematopoiesis and in skeletal development (Cosgrove,2010). Full activity of Set1/MLL methyltransferases for H3K4 methylation can only be achieved when these enzymes are assembled in a multi-protein complex. Subunits of this conserved histone methylation core complex include WDR5 (Swd3/Cps30 in yeast), RbBP5 (Swd1/Cps50 in yeast), Ash2L (Bre2/Cps60 in yeast) and Dpy30 (Sdc1/Cps25 in yeast). The WD repeat-protein 5 (WDR5), a WD40 repeat protein forming a seven bladed beta-propeller, is a key protein in this multi-protein complex and is thought to act as a scaffolding protein in the assembly of the histone methylation core complex (Zhang,2012). The catalytic Set1/MLL subunits contain a WDR5-interacting (Win) motif (LIG_WD40_WDR5_WIN_1, LIG_WD40_WDR5_WIN_2, LIG_WD40_WDR5_WIN_3) that binds to an arginine-binding pocket on WDR5 (Dharmarajan,2012). This pocket on WDR5 has also been shown to recognize unmodified, mono-, di-, and trimethylated H3K4 peptides, implying a role for WDR5 in presenting histone H3 tails for modification (Schuetz,2006). In addition, mono- and dimethylated H3K4 peptides were shown to disrupt the WDR5-MLL interaction, suggesting fine-tuned regulation of H3K4 methylation status by a complex interplay between WDR5, MLL and histone H3 (Song,2008). The WDR5 protein is also involved in recruitment of the accessory Retinoblastoma-binding protein 5 (RbBP5), which binds to the opposite side of WDR5 as the Set1/MLL subunit using a distinct motif (LIG_WD40_WDR5_VDV_1, LIG_WD40_WDR5_VDV_2) (Odho,2010). It has been shown that WDR5 is crucial for full methyltransferase activity, however only in combination with RbBP5 is this activity enhanced. Without the WDR5-RbBP5 interaction, the methyltransferase activity is weakened, leading to the assumption that WDR5 acts as a scaffold protein stabilizing the RbBP5 and MLL1 SET domain interaction with H3K4. RbBP5 is a nuclear protein that, like WDR5, belongs to a conserved family of WD repeat proteins. It contains an N-terminal beta-propeller domain that interacts with the SET domain and an unstructured acidic C-terminal tail containing a WDR5-binding motif. The interaction between RbBP5 and a catalytic SET1 protein family member cooperates with the motif-mediated interactions with WDR5 to assemble a stable and active histone methylation complex. More recently, WDR5 was found to be involved in acetylation of the histone H4 lysine 16 residue (H4K16), playing a role in the assembly of the NSL (Nonspecific lethal) complex (Dias,2014). This complex contains the catalytic MOF/KAT8, which belongs to the MYST protein family of histone acetyltransferases, and additional subunits, including WDR5, KANSL1 and KANSL2. Similar to its role in histone methylation complex assembly, WDR5 functions as a scaffold protein that binds different subunits of the NSL complex. The KANSL1 subunit conatins a Win motif (LIG_WD40_WDR5_WIN_1, LIG_WD40_WDR5_WIN_2, LIG_WD40_WDR5_WIN_3), while KANSL2 binds WDR5 via a motif similar to the WDR5-binding motif of RbBP5 (LIG_WD40_WDR5_VDV_1, LIG_WD40_WDR5_VDV_2). As the Win motifs in the methyltransferases and KANSL1 bind the same site on WDR5, and similarly, the WDR5-binding motifs in RbBP5 and KANSL2 bind the same site on WDR5, these respective interactions and the assembly of the functionally distinct complexes are mutually exclusive. |
(click table headers for sorting; Notes column: =Number of Switches, =Number of Interactions)
| Acc., Gene-, Name | Start | End | Subsequence | Logic | #Ev. | Organism | Notes |
|---|---|---|---|---|---|---|---|
| Q7Z3B3 KANSL1 KANL1_HUMAN |
590 | 596 | SDGTCVAARTRPVLSCKKRR | TP | 5 | Homo sapiens (Human) | |
| Q03164 MLL MLL1_HUMAN |
3763 | 3769 | PLNPHGSARAEVHLRKSAFD | TP | 9 | Homo sapiens (Human) | |
| O14686 MLL2 MLL2_HUMAN |
5338 | 5344 | MINPTGCARSEPKILTHYKR | TP | 6 | Homo sapiens (Human) | |
| Q8NEZ4 KMT2C KMT2C_HUMAN |
4708 | 4714 | AVNPTGCARSEPKMSAHVKR | TP | 6 | Homo sapiens (Human) | |
| Q9UMN6 KMT2B KMT2B_HUMAN |
2509 | 2515 | PLNPHGAARAEVYLRKCTFD | TP | 6 | Homo sapiens (Human) | |
| Q9UPS6 SETD1B SET1B_HUMAN |
1746 | 1752 | REHVTGCARSEGFYTIDKKD | TP | 6 | Homo sapiens (Human) | |
| O15047 SETD1A SET1A_HUMAN |
1493 | 1499 | REHQTGSARSEGYYPISKKE | TP | 6 | Homo sapiens (Human) |
Please cite:
ELM-the Eukaryotic Linear Motif resource-2024 update.
(PMID:37962385)
ELM data can be downloaded & distributed for non-commercial use according to the ELM Software License Agreement
ELM data can be downloaded & distributed for non-commercial use according to the ELM Software License Agreement