| Accession: | |
|---|---|
| Functional site class: | UEV Domain binding PTAP motif |
| Functional site description: | The PTAP motif mediates binding of several cellular and viral proteins to the UEV domain of the class E vacuolar sorting protein Tsg101. Like several other linear motifs, including the CtBP binding motif in adenovirus and USP7 binding motif in EBV, PTAP was first discovered in a viral protein. Gottlinger,1991 observed that disruption of the p6 region of HIV gag caused defective virus budding and further investigation pinpointed 4 residues, PTAP, vital to viral egress. The UEV domain of Tsg101 was not identified as the binding partner until ten years later. The solved structure of HIV gag-p6 PTAP motif bound to the UEV domain of Tsg101 showed that the motif formed one turn of a left-handed type II polyproline helix, similar to the ligands of WW and SH3 domains, while the flanking regions form extended conformations. The analysis showed that there are extensive interactions between the defined residues of the motif and the binding site, an observation consistent with the strict motif definition. |
| ELM Description: | The motif has an unusually strict definition, being fixed in all positions with the exception of the second position allowing a threonine or a serine. A randomized nonapeptide library was used for phage display and screened against the UEV domain returning 29 peptides of the consensus [AP][ST]AP allowing a non-canonical alanine at the first position. However no substitutions were tolerated in the AP region in a peptide that conferred binding. Peptide substitution analysis of the gag-p6 peptide PEPTAPPEE, confirmed that alanine was tolerated, however, proline was strongly preferred (Schlundt,2009). Nevertheless, no functional instances are known to utilize this sub-optimal variant. The UEV domain-bound motif forms one turn of a left-handed type II polyproline helix while the flanking regions form extended conformations. There are extensive intramolecular interactions between the defined residues of the motif. This observation may explain the strict motif definition. The most commonly seen variant of the peptide contains additional C-terminal prolines that have been suggested to increase affinity by stabilising the left-handed type II polyproline helix. A 9 residue peptide from the p6 chain of gag, PEPTAPPEE binds with the same affinity as full length p6, suggesting that at most the immediately flanking residues are necessary for Tsg101 binding of gag (Pornillos,2002). From the limited data available, viral proteins seem to have evolved a stronger binding affinity than host proteins. In an analysis of the TomL1 protein, HIV gag was shown to competitively out bind the UEV binding motif of TomL1 (Yanagida-Ishizaki,2008). The gag-p6 motif binds with an affinity (Kd) of 27 micromolar, however no host motifs tested (Vps37b: 55 micromolar; SCAMP3: 1975 micromolar; Tal-PSAP: 62 micromolar; and PTAP: 344 micromolar) bound with similar strength. |
| Pattern: | .P[TS]AP. |
| Pattern Probability: | 0.0001081 |
| Present in taxons: | Homo sapiens Metazoa |
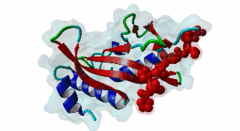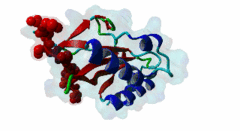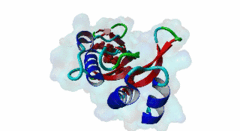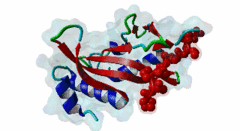
| Interaction Domain: |
UEV (PF05743)
UEV domain
(Stochiometry: 1 : 1)
|
The creation of multivesicular bodies (MVBs) is dependent on class E vacuolar protein sorting (VPS) proteins, the majority of which are part of four ESCRT (Endosomal Sorting Complexes Required for Transport) complexes (Raiborg,2009; Williams,2007). These complexes are sequentially recruited to the membranes of endosomes as cargos are sorted and targeted (although this model has been challenged (Nickerson,2007)). Tsg101 is a component of the ESCRT-1 complex, the second component of the VPS pathway (Previously ESCRT-I was the first ESCRT complex however recently HRS/STAM has been given the name ESCRT-0). Tsg101 recognises ubiquitinated cargoes via an ~145 aa N-terminal UEV domain and recruits the downstream ESCRT complexes. UEV domains are similar to E2 ubiquitin ligases, however, they have lost their ability to transfer ubiquitin due to the lack of a cysteine in the active site necessary to form thioester bonds with ubiquitin. The UEV domain also contains the PTAP-binding site and studies have shown that both ubiquitin and the PTAP motif can bind a UEV domain simultaneously. It is unknown whether UEV domains in other proteins have the ability to bind PTAP, however, the C-terminal end of Tsg101 UEV domain differs from all other known E2 folds and this difference may allow Tsg101 to specifically bind PTAP-containing peptides (Pornillos,2002). In the highly studied yeast VPS pathway, the UEV domain of the Tsg101 orthologue, Vps23, is not known to bind PTAP, suggesting that this novel E2 fold may have evolved in metazoans although this has not been shown definitively. Tsg101 itself contains a PTAP motif that has been suggested as an auto-inhibitory peptide. Analysis using liquid beta-galactosidase assays showed binding of HRS and Tsg101 increased 3-fold when the Tsg101 PTAP motif was mutated to LTAL. Several cellular proteins are known to interact with Tsg101 through PTAP motifs. However, the majority of these interactions have weak phenotypes and disruption does not abolish binding (often due to secondary interaction mediated by the C-terminal Steadiness box), although affinity is lowered. For example, mutation to the Hrs PTAP motif reduced binding by two-thirds (Lu,2003). Although several host proteins are known to interact with Tsg101 through a PTAP motif, these interactions are overshadowed by the important role that the motif plays in virus budding. The motif is essential for efficient viral particle egress of numerous enveloped RNA viruses such as the Rhabdoviridae (Vesicular Stomatitis Virus), Filoviridae (Ebola virus) and Retroviridae (HIV) (Several reviews of late domains are available such as Bieniasz,2006 and 16215227). The PTAP motif is the best known of the class of targeting motifs for the ESCRT pathway termed, rather confusingly as "late domains". "Late domains", earning the moniker of "late" due to there importance in the late stages of the viral life cycle (i.e. budding), can consist of several other motifs or groupings of motifs including; the WW domain binding motif PPxY for NEDD4, the AIP1/Alix binding motif YPxL and FPIV, and a motif of unknown binding partner in paramyxoviruses. PTAP-mediated interactions of host proteins with Tsg101 results in a diverse range of functionality. Hrs, a member of the ESCRT-0 (Hrs, STAM), associates with the early endosomes, recognizes ubiquitinated cargos and recruits the downstream ESCRT complexes through interaction with the UEV domain of Tsg101. Tsg101 mediates subsequent trafficking by targeting the cargo to the late endosome or, under certain circumstances, to be recycled to the cell surface. Two distinct regions are involved in the full affinity binding of Hrs to Tsg101; either the mutation of the PTAP motif or deletion of the C-terminal region decreased the interaction 20 fold. Tsg101/Hrs interaction mutants were unable to transport epidermal growth factor receptor (EGFR) from early to late endosome, resulting in accumulation of the EGFR in the early endosome, preventing their degradation (Lu,2003). Vps37b, a member of the ESCRT-I complex (Tsg101, Vps28, a Vps37 and a Fam125) that target cargos from the early to the late endosome and eventually to the lysosome for degradation, associates with Tsg101 through a bipartite binding interface where a coiled-coil region and a PTAP motif in Vps37b binds to the steadiness box and UEV domain in Tsg101. The dissociation constant of the interaction of the Vps37b PTAP peptide and the UEV domain was calculated as 155 micromolar. However, the PTAP motif is not the primary Tsg101 binding site of Vps37b and mutation of the PTAP binding pocket of the UEV domain did not substantially reduce the interaction between the full-length proteins (Stuchell,2004). Another ESCRT adapter protein, ALIX, can bind the proteins from both ESCRT-I and ESCRT-III and interacts with Tsg101 through a PTAP motif (Carlton,2008). Tal, a RING finger containing Tsg101-specific E3 ubiquitin ligase, is a regulator of Tsg101 functionality. Tal binds Tsg101 via one of two functional c-terminal proximal PTAP-like motifs and monoubiquitinates Tsg101 at multiple sites, deactivating it and nullifying its ability to sort cargos. Like HRS, Tal binding to Tsg101 is mediated not only by the PTAP motif but also by a second interaction, in this case a coiled-coil region of Tal interacting with the N-terminal steadyness box of Tsg101. Interestingly, the dual PTAP sites of Tal have been shown to bind with differing affinities (Amit,2004). The second PSAP motif bound more strongly of the two with a KD of 62 micromolar compared to 344 micromolar for the preceding PTAP (Schlundt,2009). Mahogunin has been shown to bind Tsg101 through a PTAP-UEV interaction and a second binding site in the region of 317-392 interacting with the steadiness box of Tsg101. Similarly to the binding of Tal to Tsg101, this interaction facilities Mahogunin`s ability to regulate Tsg101 through ubiquitination. The ability of Mahogunin to ubiquitinate Tsg101 in an in vivo ubiquitylation assay was shown to be significantly reduced by mutation of the PTAP motif to ASAA and depletion of the PTAP containing Spongiform Neurodegeneration-associated E3 Ligase Mahogunin by siRNA has been shown to slow the degradation of EGFR by disrupting transport to the Lysosome and consequently prolong the activation of EGFR downstream MAP kinase (Kim,2007). Current theory contends that Tal and Mahogunin are involved in Tsg101 recycling whereby ubiquitylation switches Tsg101 from a membrane-bound active state to an inactive soluble state. This theory would assert that an ubiquitination/de-ubiquitination cycle would disassociate cargos from Tsg101, deactivating it until a deubiquitylating enzyme (DUBs), potentially containing a PTAP-like motif, reactivates it allowing the Tsg101 to bind another cargo (Amit,2004). DUBs for this theory have yet to be identified. SCAMP3, a negative regulator of EGFR degradation by promoting recycling and redirecting endosomes back to the cell membrane, contains a functional Tsg101 binding PTAP motif (Aoh,2009). Tom1L1 contains two functional Tsg101 binding motifs and has been shown to be recruited to endosomes (Yanagida-Ishizaki,2008). Several of the Connexin family of proteins were shown to interact with Tsg101 and as the carboxyterminal tail of connexin-45 contains a PTAP motif it was suggested that this motif may mediate the interaction, however, further analyses are necessary (Auth,2009). A recent high throughput analysis of potential Tsg101 interactors suggested that polyA-binding protein 1 (PABP1), Sec24b, NF-kappaB2 and eIF4B also interact with Tsg101 through a PTAP-like motif (Schlundt,2009). Currently, no functionality or phenotype has been has been assigned to these interactions. If these interactions are biologically meaningful it would suggest an important role of the PTAP-Tsg101 interaction outside of the VPS pathway. |
(click table headers for sorting; Notes column: =Number of Switches, =Number of Interactions)
Please cite:
ELM-the Eukaryotic Linear Motif resource-2024 update.
(PMID:37962385)
ELM data can be downloaded & distributed for non-commercial use according to the ELM Software License Agreement
ELM data can be downloaded & distributed for non-commercial use according to the ELM Software License Agreement