LIG_SUFU_1
| Accession: | |
|---|---|
| Functional site class: | SUFU binding motif |
| Functional site description: | The Hedgehog (Hh) signalling pathway is involved in the regulation of essential processes in Metazoa, including embryonic development but also stem cell maintenance and tissue homeostasis in adult organisms. Defective Hh signalling results in severe phenotypic effects, ranging from developmental deficiencies to various cancers including basal cell carcinoma and medulloblastoma. Important mediators of Hh signalling are members of the GLI C2H2-type zinc-finger protein family of transcription factors. The SUFU (Suppressor of fused homolog) protein negatively regulates the Hh pathway by inhibiting the transcriptional activity of the Gli proteins, which depends on a conserved motif in the Gli transcription factors. Although there is an apparent conservation of the Hh signalling pathway in metazoans, there are some differences between Drosophila melanogaster and Mammalia with respect to the key components and the role of primary cilia in Vertebrata. |
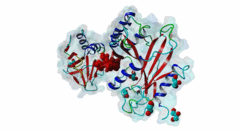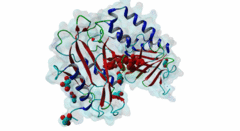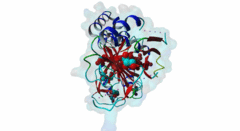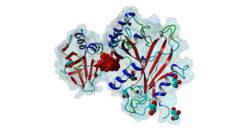
| ELM Description: | The SUFU-binding motif is found in several Gli and Gli-similar (Glis) proteins, which belong to the GLI C2H2-type zinc-finger protein family of transcription factors that mediate hedgehog signalling in Metazoa. The motif is highly conserved between Drosophila melanogaster and Homo sapiens, and is required for inhibition of Gli/Glis activity by SUFU, but in nematodes the system seems to be lost. Structural analysis shows that the motif forms a beta-strand that inserts between two beta-strands of SUFU, one from its N-terminal (PF05076) and one from its C-terminal (PF12470) domain (4BLB; 4BLD; 4KMD) (Zhang,2013; Cherry,2013). This beta-strand augmentation results in the formation of a continuous beta-sheet that spans both domains of SUFU. The first six residues of the motif form the beta-strand whose backbone forms hydrogen bonds with SUFU. In addition, the side chains of several motif residues are involved in hydrogen bonds and hydrophobic interactions with specific SUFU residues. Position 3, 4 and 5 of the motif, which are occupied by an invariant Gly, invariant His and a hydrophobic residue, respectively, form the core of the motif. The His residue in position 4 forms hydrogen bonds with Tyr147 and Asp159 of SUFU, and likely stacks with Phe155. The hydrophobic residue in position 5, most frequently Leu, binds into a hydrophobic pocket formed by Val269, Ala271 and Leu380 of SUFU. Combined mutation or deletion of these core motif residues has deleterious effects on binding and prevents inhibition of Gli activity by SUFU. The importance of these interactions for motfi binding was also shown by single or combined mutation of SUFU residues Tyr147, Phe155, Asp159 and Leu380, which has similar effects. Some other proteins contain a peptide similar to this motif, however for various reasons (see abstract below), these sequences were not yet included in the motif definition. Also, despite very high conservation of Tyr in position 2, Cys was also included as it is found in Glis3 sequences from Danio rerio. |
| Pattern: | [SV][CY]GH[LIF][LAST][GAIV]. |
| Pattern Probability: | 5.522e-08 |
| Present in taxon: | Metazoa |
| Not represented in taxon: | Nematoda |
| Interaction Domains: |
|
The hedgehog (Hh) signalling pathway regulates cell proliferation and tissue patterning during metazoan embryonic development. As proper regulation of Hh pathway is required for the correct formation of organs, defective Hh signalling results in severe developmental defects. For instance, in limbs, the Hh pathway is involved in digit development, and excess signalling leads to the formation of extra digits. In addition, aberrant activation of Hh signalling has been associated with various types of cancer, including basal cell carcinoma and medulloblastoma. The Hh pathway is conserved in vertebrates and invertebrates. However, while primary cilia do not exist in flies, Hh signalling in mammals is coupled to this organelle, which acts as a signalling centre where most of the critical components of the Hh pathway are localised and regulated (Haycraft,2005; Goetz,2010). The Hh pathway was initially identified in Drosophila melanogaster with the discovery of a gene (hh, encoding Protein hedgehog) involved in embryonic segment polarity (Nusslein-Volhard,1981; Varjosalo,2008). Hedgehog signalling is initiated by binding of the Hh ligand to its receptor Patched (Ptc) and ultimately results in the activation of several GLI C2H2-type zinc-finger protein family members (the Gli1, Gli2 and Gli3 proteins in vertebrates and the Ci protein in Drosophila melanogaster). These transcription factors can then activate the expression of specific target genes that are responsible for cell development and tissue maintenance (Varjosalo,2008). While Gli1 mainly acts as a transcriptional activator, Gli2 and Gli3 can act both as activators and repressors depending on specific post-translational modifications. For instance, in the absence of Hh signalling, the transcription factor Gli3 functions as a repressor that prevents the expression of Hh target genes, but upon Hh pathway activation differentially phosphorylated Gli3 promotes transcription (Humke,2010). The transcriptional activity of these GLI proteins is inhibited by the SUFU (Suppressor of fused homolog) protein, which thus negatively regulates Hh signalling (Ding,2000). In addition to Gli proteins, the GLI protein family also includes several other zinc-finger proteins such as the Gli-similar (Glis) and ZIC proteins. SUFU was found to also interact with and regulate Glis transcription factors. For instance, binding of SUFU to Glis3 was shown to inhibit activation of the insulin promoter by Glis3 but also stabilise Glis3 by preventing its poly-ubiquitylation-dependent proteasomal degradation (ZeRuth,2011). The interaction between SUFU and the GLI proteins it regulates is mediated by a conserved motif in these transcription factors, however, not all members of this family seem to contain a functional SUFU-binding motif. Also, while the Drosophila melanogaster family member protein Opa (P39768) contains a peptide sequence similar to the SUFU-binding motif in the Drosophila melanogaster Gli homologue Ci (P19538), the vertebrate Opa-homologous ZIC proteins do not have a sequence resembling this motif and an interaction between Opa and Sufu has not yet been characterised. In addition, several other zinc finger proteins that do not belong to the GLI family (Q8WXB4 and Q96H86) contain a similar sequence, however these peptides are not located in a disordered region and not well conserved, and thus likely not functional. |
-
Characterization of the physical interaction of Gli proteins with SUFU proteins.
Dunaeva M, Michelson P, Kogerman P, Toftgard R
J Biol Chem 2003 Feb 10; 278 (7), 5116-22
PMID: 12426310
-
Gli2 and Gli3 localize to cilia and require the intraflagellar transport protein polaris for processing and function.
Haycraft CJ, Banizs B, Aydin-Son Y, Zhang Q, Michaud EJ, Yoder BK
PLoS Genet 2005 Oct 28; 1 (4), e53
PMID: 16254602
-
Signaling from Smo to Ci/Gli: conservation and divergence of Hedgehog pathways from Drosophila to vertebrates.
Huangfu D, Anderson KV
Development 2005 Dec 12; 133 (1), 3-14
PMID: 16339192
-
Role and regulation of human tumor suppressor SUFU in Hedgehog signaling.
Cheng SY, Yue S
Adv Cancer Res 2008 Dec 05; 101 (0), 29-43
PMID: 19055941
(click table headers for sorting; Notes column: =Number of Switches, =Number of Interactions)
| Acc., Gene-, Name | Start | End | Subsequence | Logic | #Ev. | Organism | Notes |
|---|---|---|---|---|---|---|---|
| P19538 ci CI_DROME |
255 | 262 | SSAASGSYGHISATALNPMS | TP | 2 | Drosophila melanogaster (Fruit fly) | |
| Q0VGT2 Gli2 GLI2_MOUSE |
268 | 275 | SSAASGSYGHLSAGALSPAF | TP | 2 | Mus musculus (House mouse) | |
| P08151 GLI1 GLI1_HUMAN |
120 | 127 | CTSPGGSYGHLSIGTMSPSL | TP | 12 | Homo sapiens (Human) | |
| P10071 GLI3 GLI3_HUMAN |
333 | 340 | SSSASGSYGHLSASAISPAL | TP | 1 | Homo sapiens (Human) | |
| Q6XP49-2 Glis3 GLIS3_MOUSE |
348 | 355 | LSPQSEVYGHFLGVRGSCIP | TP | 3 | Mus musculus (House mouse) |
Please cite:
ELM-the Eukaryotic Linear Motif resource-2024 update.
(PMID:37962385)
ELM data can be downloaded & distributed for non-commercial use according to the ELM Software License Agreement
ELM data can be downloaded & distributed for non-commercial use according to the ELM Software License Agreement