| Accession: | |
|---|---|
| Functional site class: | PCNA binding PIP Box |
| Functional site description: | The PCNA binding motifs include the PIP Box, PIP degron, the APIM and the TLS motif. These motifs are found in proteins involved in DNA replication, repair, methylation and cell cycle control. |
| ELMs with same func. site: | LIG_PCNA_APIM_2 LIG_PCNA_PIPBox_1 LIG_PCNA_TLS_4 LIG_PCNA_yPIPBox_3 |
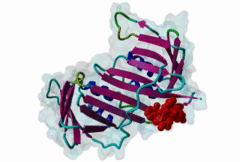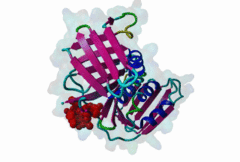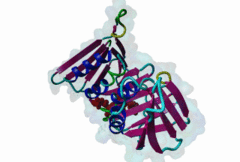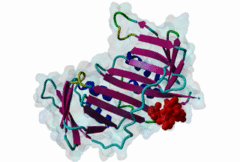
| ELM Description: | Yeast Pip Box variant. Many proteins involved in DNA replication, repair and recombination are tethered to DNA through interaction with the PCNA sliding clamp. The functional relevance of these interactions is reflected by the structural conservation of the yeast (yPCNA) and human PCNA homologs and their target protein interactions. The yeast DNA ligase I seals nicks in dsDNA during DNA replication and repair. The latter function is mediated by interaction of the DNL1 PIP Box motif with yPCNA (2OD8). As in the metazoan PIP Box (LIG_PCNA_PIPBox_1) two major binding sites are formed by the smaller ‘Q pocket’ and a large hydrophobic groove on yPCNA [Vijayakumar,2007]. The Q pocket accommodates a Gln residue, through van der Waals contacts and backbone hydrogen bonds to yPCNA Ala251 and Ala209. Compared to the metazoan motif no fungal instances bind Met, but the Q pocket accepts polar residues such as Asn in Replication factor C subunit RFC1 (1SXJ). The clamp loader (RFC 1-5)-yPCNA complex provides the only example of a full protein bound through a PIP Box motif to PCNA [Bowman,2004]. The conserved ‘hydrophobic plug’ (⏀xx⏀⏀) formed by Leu46, Phe49 and Phe51 in DNL1 adopts the canonical 310 helix that binds to the hydrophobic groove on yPCNA via van der Waals contacts and proline packing. In fungal sequences, a higher variability is observed in the second and third hydrophobic positions. Positively charged residues flanking the core motif are highly conserved in fungal instances and are included in the motif definition. As in other PIP Boxes additional interactions such as beta augmentation from DNL1 to the C-terminus of PCNA contribute to binding [Vijayakumar,2007]. Non-canonical variants include the yeast Pol η motif, which overlaps with a Rev-1 binding site (LIG_REV1ctd_RIR_1) and shows low conservation at the Q Pocket [Haracska,2001]. The SRS2 protein motif forms an ɑ-helix (3V62) instead of the canonical 310 helix and replaces the second hydrophobic position with Gln [Armstrong,2012]. |
| Pattern: | ([KR].{0,6}[QN].[^FHWY][LIVM][^P][^PFWYMLIV][FYLMWV][FYLMWVI])|([QN].[^FHWY][LIVM][^P][^PFWYMLIV][FYLMWV][FYLMWVI].{0,6}[KR]) |
| Pattern Probability: | 0.0005575 |
| Present in taxon: | Fungi |
| Interaction Domain: |
PCNA_C (PF02747)
Proliferating cell nuclear antigen, C-terminal domain
(Stochiometry: 1 : 1)
|
Eukaryotic genome duplication occurs during the DNA synthesis (S) phase of the cell cycle and ensures the transmission of genetic material to daughter cells. While this process occurs with remarkable fidelity, obstacles such as DNA lesions can lead to replication failure and chromosomes breaks, endangering genome integrity and cell viability [Moldovan,2007]. Several safeguard processes are integrated with DNA replication to sense DNA damage and allow the completion of replication using lower fidelity translesion (TLS) polymerases (Pol η, ι and κ) or initiate cell apoptosis when the damage can’t be bypassed. DNA synthesis occurs at the replication fork, where the Proliferating Cell Nuclear Antigen (PCNA) "sliding clamp" trimer acts as a scaffolding protein that orchestrates the assembly of replicative DNA polymerases, acts as a loading platform for replication factors, and integrates DNA damage and cell cycle signalling with DNA repair by recruiting TLS polymerases to the damage site to allow DNA synthesis across DNA lesions [Moldovan,2007]. The Rev1 polymerase acts as an additional molecular bridge between PCNA and TLS polymerases to facilitate polymerase exchange at stalled replication forks [Leung,2018]. Many proteins bind to PCNA through PCNA binding motifs, leading to their recruitment to the DNA replication fork. The LIG_PCNA_PIPBox_1, LIG_PCNA_APIM_2, LIG_PCNA_yPIPBox_3 and LIG_PCNA_TLS_4 motifs mediate an interaction with the PCNA PIP Box binding cleft. Among PCNA-binding proteins are enzymes involved in DNA replication, DNA repair and DNA methylation [Choe,2017]. PCNA acts as a scaffold for the integration of DNA replication with cell cycle and DNA damage signalling through the action of cell cycle regulators such as p21, which bind to PCNA using a PIP Box motif [Gulbis,1996]. The PCNA PIP-binding cleft also mediates binding of the related degron motifs DEG_CRL4_CDT2_1 and DEG_CRL4_CDT2_2. The variant PIP degron motif not only interacts with PCNA but also binds the CRL4-Cdt2 ubiquitin ligase through additional interactions, leading to the ubiquitination and proteasomal degradation of PIP degron-containing proteins following DNA damage or during S-phase [Abbas,2008]. The PIP Box, APIM and TLS variants target the same binding cleft in PCNA. The classical PCNA binding motif is termed the PIP Box [Warbrick,2000]. The core of the PIP Box motif forms a short 310 helix which interacts with a hydrophobic patch on the outer surface of the PCNA clamp through three conserved hydrophobic positions (⏀xx⏀⏀) flanked N-terminally by an additional residue which is often Q and binds to the conserved ‘Q pocket’ of PCNA (1U7B) [Bruning,2004]. PIP Boxes have a conserved hydrophobic core in vertebrate and fungal proteins, but the metazoan motif features aromatic residues at the last two positions, while fungal motifs show higher sequence variability leading to two motif variants (LIG_PCNA_PIPBox_1 and LIG_PCNA_yPIPBox_3). While the core hydrophobic positions (⏀xx⏀⏀) are shared between all PCNA motifs, additional binding determinants are present which tune the binding affinity. The APIM (LIG_PCNA_APIM_2) [Gilljam,2009] and TLS (LIG_PCNA_TLS_4) [Hishiki,2009] motif variants do not bind the Q-pocket. This is compensated by additional hydrophobic interactions in APIM motifs and by a negatively charged residue in TLS motifs that binds to human PCNA His 44, increasing binding affinity. Several non-canonical motifs exist that harbour substitutions at conserved sites, highlighting the plasticity of this binding cleft [Prestel,2019]. A prominent feature of most PIP Box motifs is the enrichment in positively charged residues in the motif core and flanking regions [Prestel,2019]. The range of binding affinities of PIP Box variants covers the low nanomolar (as seen in p21) to the micromolar range and can be strongly modulated by the presence of positive charges in the motif flanking regions, which can increase binding affinity by several orders of magnitude [Prestel,2019]. Considering the multiple processes orchestrated by PCNA, the affinity of each target might function in fine-tuning the different functional outputs of this replication hub. Post-translational modifications such as ubiquitylation and SUMOylation of PCNA act as switches that lead to the recruitment or inhibition of different binding partners and the activation of specific functional states [Moldovan,2007, Leung,2018]. For example, mono versus polyubiquitylation modulates the choice of the error-prone translesion synthesis (TLS) pathway versus the error-free template switching (TS) pathway [Leung,2018]. The mono ubiquitylation of PCNA at K164 in response to DNA damage allows the cooperative binding of ubiquitin binding motifs and PIP Box motifs present in TLS polymerases, enhancing their recruitment to the PCNA platform. During S phase, SUMOylated PCNA prevents unwanted homologous recombination. The anti-recombinogenic Srs2 helicase is recruited to SUMO-PCNA through its C-terminal domain, which contains a SUMO interaction motif (SIM) and a non-canonical PIP Box motif [Leung,2018]. The PIP Box is remarkable in that it is one of the few linear motifs found in all kingdoms of life: Fen1 has a C-terminal PIP Box-like motif in Eubacteria and Archaea as well as in Eukaryotes. Finally, PIP Box, APIM and TLS are part of a larger group of PIP-like motifs that include the TLS polymerase Rev1-interacting RIR motif (LIG_REV1ctd_RIR_1) and the mismatch repair Mlh1-interacting MIP motif (LIG_MLH1_MIPbox_1) [Ohashi,2009, Gueneau,2013]. Many PCNA targets harbour a combination of PIP Box and RIR or MIP motifs that cooperate in orchestrating DNA repair mechanisms at the PCNA hub, for example many TLS polymerases (Polη, ι and κ) contain both PCNA and Rev1 binding motifs that act cooperatively for recruiting these polymerases to PCNA. These helical motifs all have similar consensus sequences that prominently feature two adjacent aromatic residues. PIP-like motifs are functionally interlinked, as they all cooperate in different aspects of DNA repair signalling, and might show an unexpected degree of cross-functionality [Boehm,2016,Boehm,2016]. |
(click table headers for sorting; Notes column: =Number of Switches, =Number of Interactions)
Please cite:
ELM-the Eukaryotic Linear Motif resource-2024 update.
(PMID:37962385)
ELM data can be downloaded & distributed for non-commercial use according to the ELM Software License Agreement
ELM data can be downloaded & distributed for non-commercial use according to the ELM Software License Agreement