| Accession: | |
|---|---|
| Functional site class: | MDM2 binding motif |
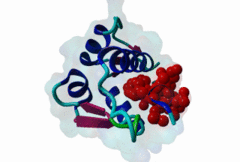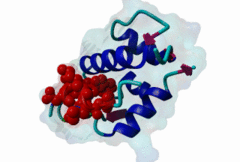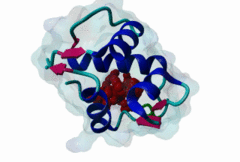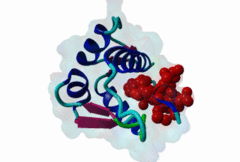
| Functional site description: | A degron motif found within the N-terminal p53 transactivation domain (TAD) (PF08563) and its relatives. The degron binds into a hydrophobic cleft in the N-terminal SWIB domain (PF02201) of the MDM2 E3 ubiquitin ligase. The sides of this pocket are formed by two helices, the bottom by two shorter helices and the ends are capped each by a three-stranded β-sheet (Kussie,1996). The p53 degron forms an amphipathic helix projecting a pair of aromatic residues deep into the MDM2 binding pocket. Regulation of p53 protein stability by Mdm2 is a key part of p53 function. |
| ELM Description: | The MDM2-binding degron motif is located in the N-terminal transactivation domain (TAD) of p53 family members, so-called BOX-I (PF08563). The motif peptide folds as an amphipathic α-helix of about 2.5 turns, which binds in the hydrophobic cleft of the MDM2 SWIB domain (Kussie,1996). The three hydrophobic amino acids Phe-19, Trp-23 and Leu-26 are all found on the same side of the p53 degron helix and are critical for binding to MDM2 since they insert deeply into the binding pocket (1YCR). For example, substitution of residues Leu-22 and Trp-23 with Gln and Ser abolishes p53-MDM2 interaction, which leads to constitutively increased p53 levels (Chehab,2000). Since the motif adopts the α-helical fold when bound, proline residues are excluded from the non-conserved positions in the motif. When looking at p53 sequence alignments, there is sometimes an extra non-conserved residue between the Trp and Leu residues: This is likely to be due to underwinding of the end turn of the helix. |
| Pattern: | F[^P]{3}W[^P]{2,3}[VIL] |
| Pattern Probability: | 0.0000212 |
| Present in taxon: | Metazoa |
| Interaction Domain: |
SWIB (PF02201)
SWIB/MDM2 domain
(Stochiometry: 1 : 1)
|
The p53 protein (P04637) has a role in restricting genome mutation, being nicknamed the “guardian of the genome”. In response to DNA damage, p53 is upregulated and acts as a transcription factor influencing expression of genes involved in cell cycle control. p53 can control gene expression by its N-terminal transactivation domain (TAD, PF08563). In humans, p53 is encoded by the TP53 gene, which is described as a tumour suppressor gene with a high frequency of mutation in cancer (Oliver,2011). The anti-carcinogenic function of p53 is achieved by influencing genomic stability, angiogenesis, and apoptosis. Further, p53 influences several cellular processes, including metabolism and in the cancer context, invasion and metastasis, as well as communication within the tumour environment (Bieging,2014). p53 induction is triggered by DNA double strand breaks or collapsed DNA replication forks, activating the cell cycle checkpoint kinases ATM (Q13315) and ATR (Q13535), respectively. These kinases can directly phosphorylate p53 but are also responsible for activation of other kinases, which may then phosphorylate p53 on other sites. Phosphorylation is not only important for p53 stabilization but also for its interaction with transcriptional co-factors. This is crucial for activation of p53 target genes and for protective responses such as cell cycle arrest, DNA repair, apoptosis, and senescence (Bieging,2014). The E3 ubiquitin-protein ligase MDM2 (Murine double minute 2, Q00987) is one of the principal p53 modulators by mediating its ubiquitination and thereby targeting p53 for proteasomal degradation (Wade,2010). To accomplish this, MDM2 uses a dual-site mechanism involving interactions between its N-terminal hydrophobic pocket in the SWIB domain (PF02201) and the natively disordered transactivation domain (TAD) of p53, so-called BOX-I. Subsequently, binding of the acidic domain to an ubiquitination signal in the central DNA binding domain (DBD, PF00870) of p53 is promoted by a conformational change of MDM2 (Wallace,2006). MDM2 is located both in the nucleus and in the cytoplasm (Oliver,2011). Inactivation of p53 is the most prevalent defect in human cancers. MDM2 can show increased expression levels in certain tumours such as retinoblastoma, where the p53 protein is not mutated, acting to impair p53 function. This compromises the ability of p53 to act as a tumour suppressor by inhibiting cell cycle arrest (Vousden,2007). Upregulation of MDM2 can occur by gene duplication and in tumour cells binding of MDM2 at TAD of p53 will both inhibit its transactivation function and cause proteasome-mediated degradation by ubiquitination (Bieging,2014). Inhibition of p53 binding to MDM2 can recover p53 activity in tumour cells and is therefore being evaluated as an approach to cancer therapy. For this purpose, analogue peptides and chemical ligands (so called “nutlins”) with high affinity to MDM2 have been developed. These are capable of disrupting interactions between WT p53 and MDM2, preventing p53 degradation (Brown,2009). However, in healthy cells MDM2 is a p53 responsive gene since transcription is induced when p53 is stabilized upon DNA damage (Fu,2010). MDM4 (also MDMX, O15151) is another negative regulator of p53. It shows structural similarity to MDM2 and binds also to the same degron of p53. This inhibits transactivation function of p53 but does not trigger p53 degradation (Bieging,2014). MDM2-p53 interaction is perturbed by p53 phosphorylation (see above), as well as by activation of the ARF tumour suppressor. ARF is stimulated by the E2F transcription factor, which is liberated due to hyperproliferative signals. ARF is able to antagonize ubiquitin ligase activity, and/or transferring MDM2 to nucleoli. Thus, ARF activation contributes to p53 stability and activity (Bieging,2014). The transcription factors p63 and p73 belong to the p53 family and show structural similarity. Like p53, both proteins have a disordered transactivation domain containing the motif that binds to the hydrophobic cleft of MDM2 and can mediate the transactivation of p53-responsive genes. However, MDM2 binding affinity of p53 is twice as high as the affinity of p73. p63 binding to MDM2 is reported to be 20 times weaker than p53 binding (Shin,2015). MDM2 binding abolishes transactivation function of p73, but in contrast to p53 it does not induce ubiquitin-mediated proteosomal degradation (Balint,1999). The cell fate determinant NUMB (P49757) was identified as a MDM2 binding partner that employs a dual-site docking mechanism (Juven-Gershon,1998). It was postulated that NUMB mediates binding only to the PTB domain, which interacts with the N-terminal hydrophobic pocket and the acidic domain of MDM2 (Sczaniecka,2012). But we note the presence of the MDM2 degron motif in NUMB, which is conserved in the paraloguous protein NUMBL (NUMB-like protein, Q9Y6R0), suggesting that these two proteins also have a two-sited binding mechanism like p53. NUMB plays an important role in the Notch signaling pathway by acting as a Notch antagonist. It promotes degradation of Notch by interacting with its active intracellular domain. Thus, NUMB prohibits its localization to the nucleus and its activity as a transcription factor (Gulino,2010). As a ligand of MDM2, NUMB prevents p53 degradation by disrupting the MDM2-p53 complex. The MDM2-mediated p53 ubiquitination is prohibited and p53 level stabilized (Sczaniecka,2012). |
(click table headers for sorting; Notes column: =Number of Switches, =Number of Interactions)
| Acc., Gene-, Name | Start | End | Subsequence | Logic | #Ev. | Organism | Notes |
|---|---|---|---|---|---|---|---|
| Q9Y6R0 NUMBL NUMBL_HUMAN |
577 | 584 | APELDPFEAQWAALEGKATV | TP | 1 | Homo sapiens (Human) | |
| P49757 NUMB NUMB_HUMAN |
616 | 623 | TCPVDPFEAQWAALENKSKQ | TP | 11 | Homo sapiens (Human) | |
| O15350 TP73 P73_HUMAN |
15 | 22 | PDGGTTFEHLWSSLEPDSTY | TP | 4 | Homo sapiens (Human) | |
| Q9H3D4 TP63 P63_HUMAN |
55 | 62 | FLSPEVFQHIWDFLEQPICS | TP | 3 | Homo sapiens (Human) | |
| P04637 TP53 P53_HUMAN |
19 | 26 | PLSQETFSDLWKLLPENNVL | TP | 4 | Homo sapiens (Human) |
Please cite:
ELM-the Eukaryotic Linear Motif resource-2024 update.
(PMID:37962385)
ELM data can be downloaded & distributed for non-commercial use according to the ELM Software License Agreement
ELM data can be downloaded & distributed for non-commercial use according to the ELM Software License Agreement