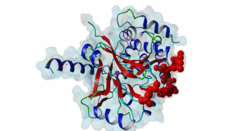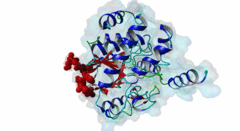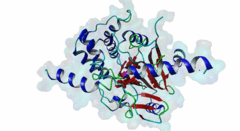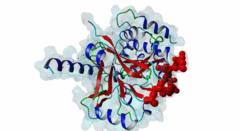
| Accession: | |
|---|---|
| Functional site class: | PP1-docking motif SILK |
| Functional site description: | Protein phosphatase 1 (PP1) is a ubiquitous serine/threonine protein phosphatase that is highly conserved throughout eukaryotic cells. It catalyzes the majority of protein dephosphorylation events and is involved in regulating essential cellular functions including carbohydrate metabolism, muscle contraction, entry into mitosis and RNA splicing (Bollen,2010; Huang,1999). The PP1 apoenzyme is a single catalytic domain that can interact with more than 200 regulators, converting it into hundreds of highly specific holoenzymes. The catalytic site of PP1 is at the intersection of three potential substrate-binding regions: the acidic, hydrophobic and C-terminal grooves (Peti,2013). Most regulatory proteins interact with PP1 at the catalytic site via the RVXF docking motif (DOC_PP1_RVXF_1) but docking motifs such as the SILK motif (DOC_PP1_SILK_1) and the MyPhoNE motif also play essential roles in regulating PP1 activity and substrate specificity (Hendrickx,2009). |
| ELMs with same tags: |
|
| ELM Description: | The SILK motif is embedded in multiple proteins and essential for docking regulatory proteins to PP1c. It generally cooperates with and occurs N-terminal to an RVXF motif (DOC_PP1_RVXF_1), and is very conserved throughout evolution (Hendrickx,2009). Interaction with PP1c involves hydrophobic associations and hydrogen bonds. In the first position of the motif mainly charged residues are found. They provide a basis for hydrogen bonds but are not specific. The second position requires a glycine or serine residue due to steric restrictions. Serine additionally provides hydrogen bonds with PP1c. The third position is always occupied by isoleucine, which is buried in a hydrophobic pocket formed by the residues Leu55, Phe119 and Leu59 of PP1c. The invariant leucine residue in position 4 is essential for the specificity of the interaction and is buried in a pocket on PP1c formed by Pro50, Leu53, and Phe119. In over 90% of the cases, leucine is followed by a lysine, which hydrogen bonds with Asp166 in PP1c. An arginine in this position is also possible. The last position of the motif often, but not always, contains an asparagine, which forms hydrogen bonds with the PP1c main chain carbonyl oxygen and main chain nitrogen of the residues 52 and 54. Acidic residues were never observed in this position in multiple sequence alignments of the validated instances and homologues thereof, suggesting the possibility of regulation of motif binding by phosphorylation. It is suggested that the role of PP1c Glu54 and Glu56 is to maintain a negatively charged surface that attracts and orientates the lysine/arginine of the SILK motif, thereby promoting interactions involving the isoleucine and leucine (2O8G) (Hurley,2007). Deletion of the SILK motif causes a large reduction in the inhibitory function of IPP-2 (Connor,2000; Huang,1999) and it is necessary for stable complex assembly between PP1, Spinophilin and IPP-2 (Dancheck,2011). |
| Pattern: | .[GS]IL[KR][^DE] |
| Pattern Probability: | 0.0000378 |
| Present in taxon: | Eukaryota |
| Interaction Domain: |
Metallophos (PF00149)
Calcineurin-like phosphoesterase
(Stochiometry: 1 : 1)
|
The ubiquitous serine ⁄ threonine protein phosphatases of type 1 (PP1) and type 2A (PP2A) belong to the phosphoprotein phosphatase (PPP) superfamily (Peti,2013). Together, they are responsible for more than 90 % of the protein phosphatase activity in eukaryotic cells. PP1 regulates diverse cellular processes such as cell cycle progression, apoptosis, protein synthesis, muscle contraction, glycogen metabolism, transcription and neuronal signalling (Hurley,2007; Aggen,2000). Three different genes in the mammalian genome encode four distinct catalytic subunits PP1α, PP1β⁄δ and the splice variants PP1γ1 and PP1γ2. These isoforms show a sequence identity of about 90% and most of the amino acid differences are found in the N- and C-terminal extremities. While Saccharomyces cerevisiae with only one PP1 gene (Glc7) is an exception, many Bacteria759 have multiple PP1 genes: eight are found in Arabidopsis thaliana, four in Drosophila melanogaster and 30 are predicted in Caenorhabditis elegans (Moorhead,2007, Wakula,2003). With more than 200 target proteins the catalytic subunit (PP1c) lacks substrate specificity (Bollen,2010). It is recruited by the regulatory subunits to dephosphorylate phosphothreonine or phosphoserine residues in the target substrates. Using one or more of these regulators, PP1c is converted into hundreds of very specific holoenzymes. The catalytic site of PP1c is located at the intersection of three substrate-binding grooves: the acidic, the hydrophobic and the C-terminal binding groove (Terrak,2004). It is surrounded by acidic residues and contains two metal ions. In mammalian tissues these ions are Fe2+ and Zn2+, whereas in bacteria Mn2+ is incorporated (Bollen,2010). PP1-interacting proteins (PIPs) function as targeting subunits, inhibitors and substrates. More than half of all PIPs inhibit PP1 when glycogenphosphorylase is used as a substrate. The IPP-1 (Protein phosphatase inhibitor 1, Q13522) protein and its homolog DARPP-32 (Dopamine- and cAMP-regulated phosphoprotein, Q9UD71) bind to and block the PP1 catalytic site after being phosphorylated on a specific threonine residue, thus acting as pseudosubstrates, while IPP-2 (Protein phosphatase inhibitor 2, P41236) does not require prior phosphorylation for the formation of a stable inactive complex with PP1c (Huang,1999; Connor,2000). PIPs are predicted to contain regions that are intrinsically disordered, an attribute that favours their binding to a large surface area of PP1 via multiple docking motifs. Most of the known PP1-docking motifs are short, highly conserved and their flanking regions contain less conserved segments (Hendrickx,2009). They play an essential role in the regulation of PP1 activity and substrate specificity. Generally, PIPs contain multiple distinct PP1-docking motifs, which likely mediate cooperative binding to PP1. The RVXF motif (DOC_PP1_RVXF_1) is present in more than 90% of all known PP1 regulators. It functions primarily as an anchoring motif, as it does not influence the structure of PP1. The RVXF binding site is ~20 Å away from the PP1c active site. The hydrophobic groove of PP1c interacts with the RVXF motif in regulatory proteins, binding the peptide backbone by beta-strand augmentation (Peti,2013; Moorhead,2007). The SILK motif (DOC_PP1_SILK_1) is highly conserved among Opisthokonta and Amoebozoa and occurs in IPP-2 and several other vertebrate PP1 interactors. It binds in the hydrophobic groove at the back side of PP1 and is located N-terminal to the RVXF motif in proteins where these two motifs co-occur (Dancheck,2011; Hendrickx,2009). Similar to the RVXF motif, the SILK motif acts as an anchoring site and does not change the conformation of PP1. The myosin phosphatase N-terminal element or MyPhoNE motif contributes to substrate selection and is present in MYPT1 (Myosin phosphatase target subunit 1, O14974) and a few other PIPs. It is also located N-terminal to the RVXF sequence when both are present, lying within a five-turn α-helix that faces hydrophobic residues in a shallow hydrophobic cleft on PP1 (Bollen,2010). |
(click table headers for sorting; Notes column: =Number of Switches, =Number of Interactions)
Please cite:
ELM-the Eukaryotic Linear Motif resource-2024 update.
(PMID:37962385)
ELM data can be downloaded & distributed for non-commercial use according to the ELM Software License Agreement
ELM data can be downloaded & distributed for non-commercial use according to the ELM Software License Agreement